research groups
Metalloproteins
summary

- Vila, Alejandro Location: CCT
Email: vila@ibr-conicet.gov.ar
- Hita, Francisco
RESEARCH LINES
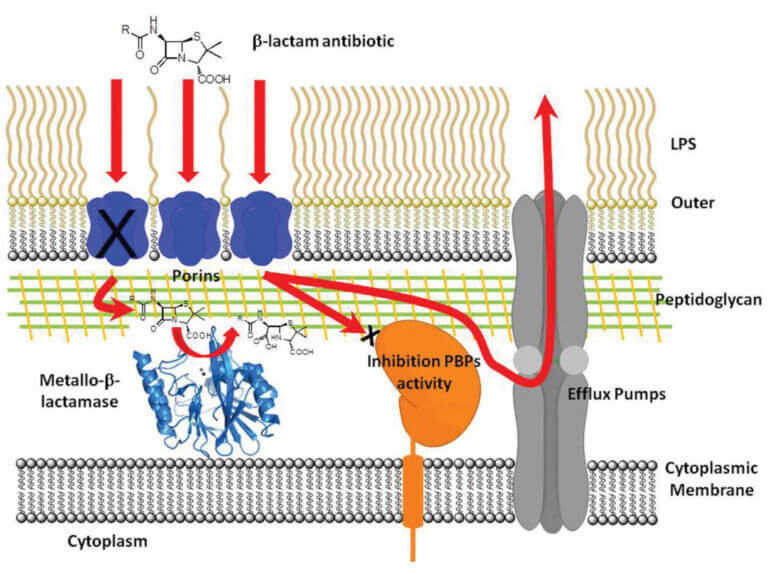
Antibiotic resistance: Evolution, structural diversity and catalytic mechanism of metallo-β-lactamases
β-Lactam antibiotics are the most widely used chemotherapeutic agents for the treatment of bacterial infections. The main mechanism of resistance to these drugs is the synthesis of β-lactamases, enzymes that hydrolyze and inhibit the action of these antibiotics. Metallo-β-lactamases (MβLs) constitute the most recent generation of these enzymes. MβLs have a broad spectrum of action, inactivating all β-lactam antibiotics, including carbapenems (the last resort used in clinical practice).
The dissemination of genes encoding these enzymes in opportunistic and pathogenic organisms is now global. This situation is exacerbated by the structural diversity of the different MβLs, which makes it difficult to design an efficient inhibitor for these enzymes. The NDM-1 enzyme (http://www.bbc.co.uk/news/health-10925411), in particular, is rapidly spreading worldwide, and there are no clinically used inhibitors for these enzymes, posing a threat to global health.
Our goal is to elucidate the structure-function relationship of these enzymes through biochemical, structural, mechanistic, and evolutionary studies, with the ultimate goal of designing a clinically applicable inhibitor. To date, we have succeeded in: (1) proposing a common catalytic mechanism for MβLs by characterizing a reaction intermediate, (2) identifying their functional in vivo species, and (3) exploring their potential evolutionary mechanisms. We are currently using this prior knowledge to design and generate MβL inhibitors, under the hypothesis that, despite their structural diversity, MβLs act using the same catalytic mechanism.
Our group conducts an interdisciplinary study, using techniques from molecular biology, biochemistry, structural biology, and modern enzymology. Mechanistic studies are performed using rapid mixing techniques and various spectroscopies to follow changes in the active site during catalytic turnover on the millisecond timescale, with the goal of identifying reaction intermediates.
We use crystallography and/or Nuclear Magnetic Resonance (NMR) to characterize the binding mode of potential inhibitors to these enzymes. We also use in vitro directed evolution strategies as a means to predict the evolution of these enzymes in hospital environments and to understand how mutations are fixed in the evolutionary process.
Copper in biological systems: Control of reactivity and insertion mechanism of copper electron transfer sites
La respiración celular depende del control fino de una serie de procesos de oxido-reducción, que culminan en la reducción de oxígeno llevada a cabo por las citocromo c oxidasas. El cobre es un metal de transición esencial para la transferencia electrónica en estas enzimas, siendo a la vez tóxico a altas concentraciones. Por lo tanto, su concentración intracelular y mecanismos de transporte y almacenamiento deben estar finamente regulados.
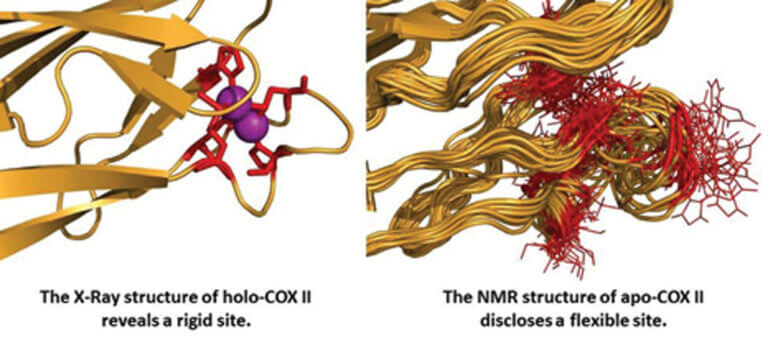
Our group studies: (1) the biological mechanisms of copper ion transport and insertion into terminal oxidases and (2) the regulation of the function of these sites by fine-tuning their electronic and three-dimensional structure. We have identified the proteins involved in copper transport and insertion into the CuA site of bacterial cytochrome oxidases and structurally characterized the metal-binding mechanism using NMR. To study the electronic structure of these sites, classical structural biology methods are insufficient, as they only provide the three-dimensional structure.
In our laboratory, we use several spectroscopic techniques (mainly NMR) to analyze the electron distribution at copper sites and study how it determines long-distance electron transfer. We have used this strategy to describe the fine-tuning of reactivity in blue copper proteins, and we are currently applying our experience to the study of the CuA copper site in cytochrome c oxidases. We have proposed that the high efficiency of the CuA site in long-distance electron transfer is primarily due to the presence of low-energy excited electronic states that are populated at room temperature.
Images of our research lines