research groups
Biophysics of Molecular Recognition
summary

- Rasia, Rodolfo Location: CCT
Email: rasia@ibr-conicet.gov.ar
- Cenizo, Zoe
RESEARCH LINES
Induced protein folding in gene regulation in plants
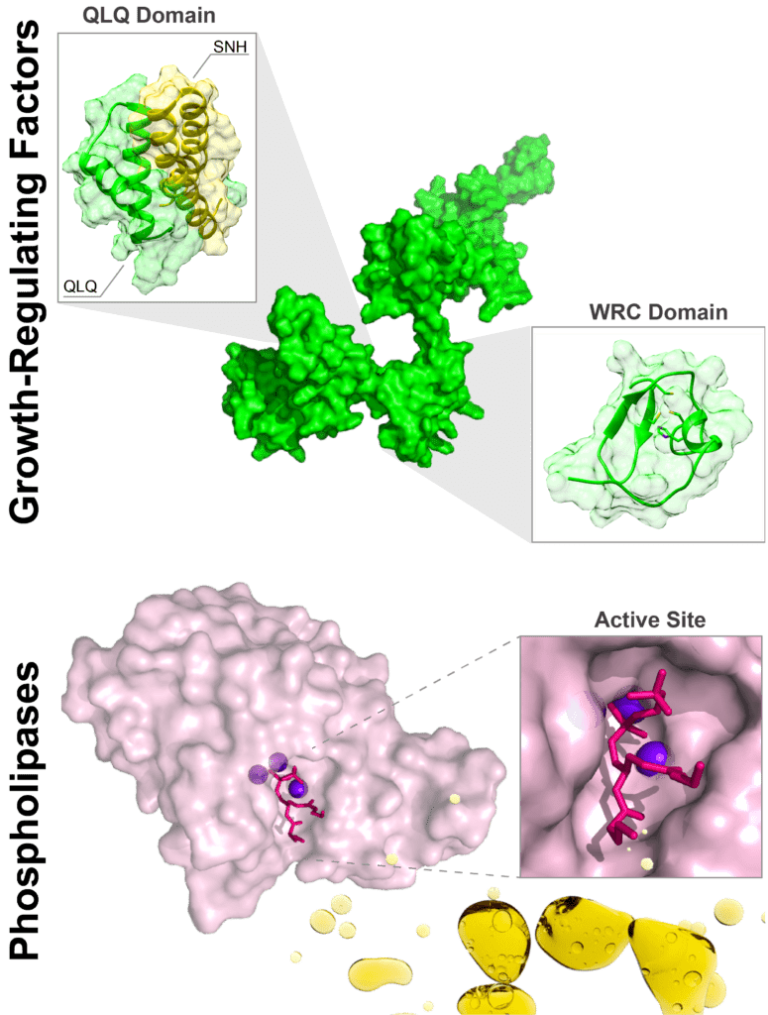
Gene expression in eukaryotic organisms is finely regulated through the action of different proteins. Transcription factors (TFs) recognize DNA sequences and thus control the transcription of adjacent genes. TFs are in turn aided by transcriptional coactivators (TCs), which contribute to the reorganization of chromatin. In this project we study the GRF:GIF system of TFs:TCs in plants. The GRF family of TFs has 9 members and the GIF family of TCs has 3. The proteins in these two families are mostly disordered. GRF TFs recognize DNA through their WRC domains. In turn, the interaction between the proteins of these two families is mediated by the QLQ domains, in the GRF TFs , and the SNH domains, in the GIF TCs.
The aims of the project are to study:
-The formation of complexes mediated by the QLQ and SNH domains. Both domains are disordered in their free form, and their interaction occurs through an unusual mechanism of mutual synergistic folding.
-The structure of the WRC domains and their mode of recognition of specific DNA sequences.
-The presence of molecular recognition elements in the extensive disordered regions of the protein and their participation in the regulation of the global conformation of the complexes.
Enzymes optimization
The use of enzymes as catalysts is prevalent in different modern industrial processes, where their specificity and mild reaction conditions are extremely valuable. The reaction media used in these settings differ substantially from the cellular or extracellular environment in which enzymes have naturally evolved. The efficient use of enzymes in these hostile environments has required, in many cases, their modification through protein engineering, leading to better adapted variants that maintain their functionality.
Our projects seek to obtain variants of enzymes with greater stability and activity than those obtained from natural sources. We use bacterial phospholipases C as model proteins. These enzymes are used in the industrial process of enzymatic oil degumming, with significant economic importance in the Greater Rosario region. We use combination of known sequences and machine learning methods to generate new protein sequences. We then evaluate the stability and activity of selected variants using spectroscopic and biochemical techniques. The analysis of the results that we obtain in these studies allows us to address unresolved fundamental questions on protein stability and reactivity.
Signal integration in the control of plant cell growth and early development
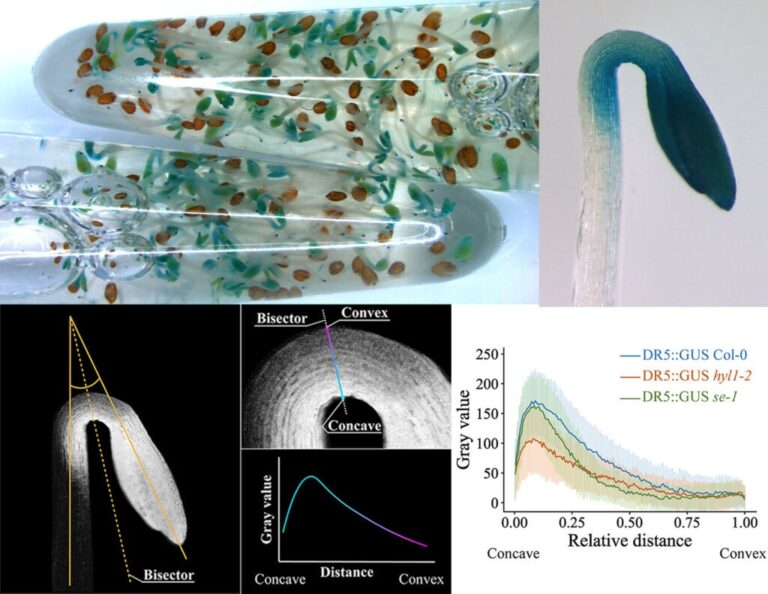
Plants have developed sophisticated molecular mechanisms to perceive and integrate environmental and endogenous signals in order to elaborate a proper response for growth and development. We use the model plant Arabidopsis thaliana to study early development, since seeds germinate and grow in the darkness (skotomorphogenesis) until the seedlings reach the light to synthesize and activate the photosynthetic machinery (photomorphogenesis) in order to change to an autotrophic growth. The research framework for this project has three working packages that tackle how plants integrate environmental and endogenous signals to decide and execute cell growth and developmental programs:
1-Molecular mechanisms controlling early plant development. Convergence of light/darkness, biogenesis of microRNAs and auxin signals for a proper skotomorphogenic development.
2-Functional and mechanistic conservation of the master regulator HY5. We study the conservation of HY5 transcription factor activity between Arabidopsis and the model tree poplar, which is essential for biomass production to confront climate change.
3-Cellular architecture in the control of plant cell growth. We study the functional interdependence between cortical microtubules, necessary for cellular growth, light/dark signals and the phytohormones auxins.
Images of our research lines
PUBLICATIONS AND PATENTS
Characterization of a novel thermostable phospholipase C from T. kodakarensis suitable for oil degumming.
Characterization of a novel thermostable phospholipase C from T. kodakarensis suitable for oil degumming.
HYPONASTIC LEAVES 1 is required for proper establishment of auxin gradient in apical hooks.
HYPONASTIC LEAVES 1 is required for proper establishment of auxin gradient in apical hooks.
Dual function of HYPONASTIC LEAVES 1 during early skotomorphogenic growth in Arabidopsis.
Dual function of HYPONASTIC LEAVES 1 during early skotomorphogenic growth in Arabidopsis.
Conformational sampling of the intrinsically disordered dsRBD-1 domain from Arabidopsis thaliana DCL1.
Conformational sampling of the intrinsically disordered dsRBD-1 domain from Arabidopsis thaliana DCL1.
The key role of electrostatic interactions in the induced folding in RNA recognition by DCL1-A.
The key role of electrostatic interactions in the induced folding in RNA recognition by DCL1-A.
Sede CCT Rosario
Ocampo y Esmeralda, Predio CONICET-Rosario
2000 Rosario, Santa Fe, Argentina
Tel. 54-341-4237070 / 4237500 / 4237200
Sede Facultad de Ciencias Bioquímicas y Farmacéuticas
Universidad Nacional de Rosario - Suipacha 531
2000 Rosario, Santa Fe, Argentina
Tel. +54 341 4350596 / 4350661 / 4351235
🔬 El IBR suma 9 proyectos seleccionados en Investigación Orientada 2025 de @ProduccionSF y @CienciaSantaFe.
Biotecnología, salud y sostenibilidad para fortalecer el vínculo entre ciencia, innovación y desarrollo territorial.

